Note
Go to the end to download the full example code
DCIP with PyGIMLi (Synthetic example)#
If you are running this notebook locally, make sure you’ve followed steps here to set up the environment. (This environment.yml file specifies a list of packages required to run the notebooks)
Using the DCIP (Direct Current, Induced Polarization) solver provided by
PyGIMLi, we use different cofi
solvers to solve the corresponding inverse problem.
Note: This notebook is adapted from a PyGIMLi example: Naive complex-valued electrical inversion
The key difference between ERT and DCIP as implemented in PyGIMLi is that for DCIP resistivties are expressed as complex numbers with the real part representing the resistivity and the phase angle presenting the chargeability. This means that entries into the model vector and the data vector are complex numbers and that DCIP inversions using PyGIMLI rely on the induced polarization field measurements being expressed in the frequency domain.
While numpy.linalg.solve is able to call the appropriate Lapack
subroutine for a complex linear system cgesv or zcgesv, other
solvers typically expect the model vector and data vector to be real.
This means that the complex system of equation needs to be transformed
into a real system of equations. Such a transformation needs to be
accounted for in the user provided functions for the objective function,
Hessian and gradient; care must also be taken when transforming the data
covariance matrix.
The linear equation $ A x =b $ with the elements of \(A\), \(b\) and \(x\) being complex numbers can be rewritten using real numbers as follows
with \(b=( b_1^r+b_1^c i, b_2^r+b_2^c i,...,b_n^r+b_n^c i)\) being rewritten as \((b^r,b^c)\) with \(b^r=(b_1^r,b_2^r,...,b_n^r)\) and \(b^c=(b_1^c,b_2^c,...,b_n^c)\) and analogus reordering for \(A\) and \(x\).
See https://ijpam.eu/contents/2012-76-1/11/11.pdf for more details.
1. Import modules#
# -------------------------------------------------------- #
# #
# Uncomment below to set up environment on "colab" #
# #
# -------------------------------------------------------- #
# !pip install -U cofi
# !pip install -q condacolab
# import condacolab
# condacolab.install()
# !mamba install -c gimli pygimli=1.3
We will need the following packages:
numpyfor matrices and matrix-related functionsmatplotlibfor plottingpygimlifor forward modelling of the problemcofifor accessing different inference solvers
import numpy as np
import matplotlib.pyplot as plt
import pygimli
import cofi
np.random.seed(42)
Below we define a set of utility functions that help define the problem, generating data and making plots. Feel free to skip reading the details of these utility functions and come back later if you want.
1.1. Helper functions for complex numbers#
def rho_phi_to_complex(rho, phi): # rho * e^(phi * i)
return pygimli.utils.toComplex(rho, phi)
def rho_phi_from_complex(complx): # |complx|, arctan(complx.imag, complx.real)
return np.abs(complx), np.arctan2(complx.imag, complx.real)
def complex_to_real(complx): # complx vector of size n -> size 2n
return pygimli.utils.squeezeComplex(complx)
def complex_from_real(real): # real vector of size n -> size n/2
return pygimli.utils.toComplex(real)
1.2. Helper functions for PyGIMLi modelling#
# Utility Functions
x_inv_start = -2
x_inv_stop = 52
y_inv_start = -20
y_inv_stop = 0
def survey_scheme(start=0, stop=50, num=51, schemeName="dd"):
scheme = pygimli.physics.ert.createData(elecs=np.linspace(start=start, stop=stop, num=num),schemeName=schemeName)
return scheme
def model_true(
scheme,
start=[-55, 0],
end=[105, -80],
anomalies_pos=[[10,-7],[40,-7]],
anomalies_rad=[5,5],
rhomap=[[1, rho_phi_to_complex(100, 0 / 1000)],
# Magnitude: 50 ohm m, Phase: -50 mrad
[2, rho_phi_to_complex(50, 0 / 1000)],
[3, rho_phi_to_complex(100, -50 / 1000)],]
):
world = pygimli.meshtools.createWorld(start=start, end=end, worldMarker=True)
for s in scheme.sensors(): # local refinement
world.createNode(s + [0.0, -0.1])
geom = world
for i, (pos, rad) in enumerate(zip(anomalies_pos, anomalies_rad)):
anomaly = pygimli.meshtools.createCircle(pos=pos, radius=rad, marker=i+2)
geom += anomaly
mesh = pygimli.meshtools.createMesh(geom, quality=33)
return mesh, rhomap
def ert_simulate(mesh, scheme, rhomap, noise_level=1, noise_abs=1e-6):
pg_data = pygimli.physics.ert.simulate(mesh, scheme=scheme, res=rhomap, noiseLevel=noise_level,
noise_abs=noise_abs, seed=42)
# data.remove(data["rhoa"] < 0)
data_complex = rho_phi_to_complex(pg_data["rhoa"].array(), pg_data["phia"].array())
data_log_complex = np.log(data_complex)
return pg_data, data_complex, data_log_complex
def ert_manager(pg_data, verbose=False):
return pygimli.physics.ert.ERTManager(pg_data, verbose=verbose, useBert=True)
def inversion_mesh(ert_mgr):
inv_mesh = ert_mgr.createMesh(ert_mgr.data)
# print("model size", inv_mesh.cellCount()) # 1031
ert_mgr.setMesh(inv_mesh)
return inv_mesh
def ert_forward_operator(ert_mgr, pg_data, inv_mesh):
forward_oprt = ert_mgr.fop
forward_oprt.setComplex(True)
forward_oprt.setData(pg_data)
forward_oprt.setMesh(inv_mesh, ignoreRegionManager=True)
return forward_oprt
def reg_matrix(forward_oprt):
region_manager = forward_oprt.regionManager()
region_manager.setConstraintType(2)
Wm = pygimli.matrix.SparseMapMatrix()
region_manager.fillConstraints(Wm)
Wm = pygimli.utils.sparseMatrix2coo(Wm)
return Wm
def starting_model(data, inv_mesh, rho_val=None, phi_val=None):
rho_start = np.median(data["rhoa"]) if rho_val is None else rho_val
phi_start = np.median(data["phia"]) if phi_val is None else phi_val
start_model_val = rho_phi_to_complex(rho_start, phi_start)
start_model_complex = np.ones(inv_mesh.cellCount()) * start_model_val
start_model_log_complex = np.log(start_model_complex)
start_model_log_real = complex_to_real(start_model_log_complex)
return start_model_complex, start_model_log_complex, start_model_log_real
def model_vector(rhomap, mesh):
return pygimli.solver.parseArgToArray(rhomap, mesh.cellCount(), mesh).array()
1.3. Helper functions for plotting#
def plot_model(mesh, model_complex, title):
rho, phi = rho_phi_from_complex(model_complex)
fig, axes = plt.subplots(1,2,figsize=(10,3))
pygimli.show(mesh, data=rho, label=r"$\Omega m$", ax=axes[0])
axes[0].set_xlim(x_inv_start, x_inv_stop)
axes[0].set_ylim(y_inv_start, y_inv_stop)
axes[0].set_title("Resistivity")
pygimli.show(mesh, data=phi * 1000, label=r"mrad", ax=axes[1])
axes[1].set_xlim(x_inv_start, x_inv_stop)
axes[1].set_ylim(y_inv_start, y_inv_stop)
axes[1].set_title("Chargeability")
fig.suptitle(title)
def plot_data(pg_data, data_complex, title):
rho, phi = rho_phi_from_complex(data_complex)
fig, axes = plt.subplots(1,2,figsize=(10,4))
pygimli.physics.ert.showERTData(pg_data, vals=rho, label=r"$\Omega$m", ax=axes[0])
axes[0].set_title("Apparent Resistivity")
pygimli.physics.ert.showERTData(pg_data, vals=phi*1000, label=r"mrad", ax=axes[1])
axes[1].set_title("Apparent Chargeability")
fig.suptitle(title)
2. Define the problem#
We first define the true model, the survey and map it on a computational mesh designed for the survey and true anomaly.
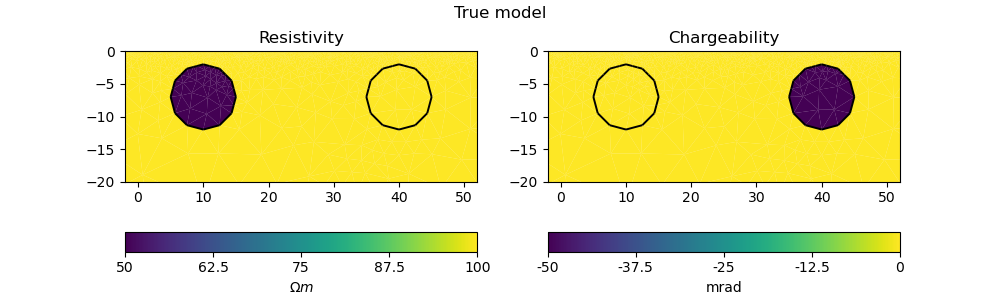
2.1. True model#
# PyGIMLi - define measuring scheme, geometry, forward mesh and true model
scheme = survey_scheme()
mesh, rhomap = model_true(scheme)
# plot the true model
plot_model(mesh, model_vector(rhomap, mesh), "True model")
2.2. Generate synthetic data#
Generate the synthetic data as a container with all the necessary information for plotting:
pg_data, data_complex, data_log_complex = ert_simulate(mesh, scheme, rhomap)
plot_data(pg_data, data_complex, "(Synthetic) Data Observatons")
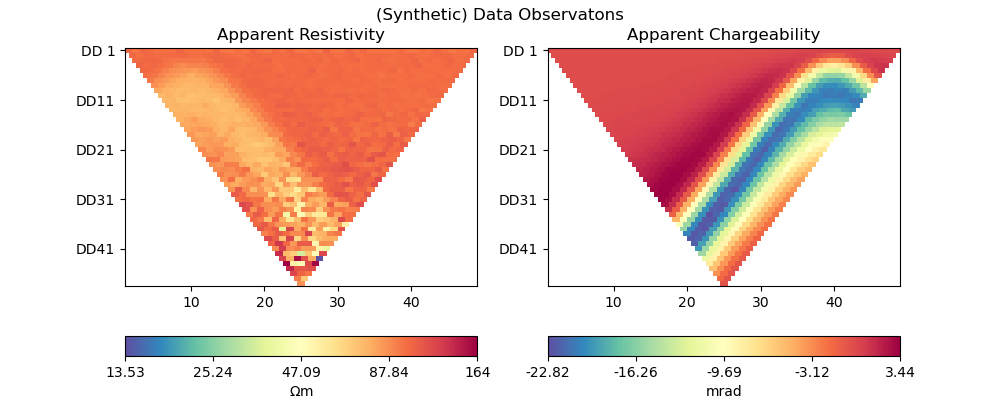
relativeError set to a value > 0.5 .. assuming this is a percentage Error level dividing them by 100
Data IP abs error estimate (min:max) 7.191825579044176e-10 : 0.0002250043443897723
2.3. ERTManager#
Further, we create a pygimli.ert.ERTManager instance to keep record
of problem-specific information like the inversion mesh, and to perform
forward operation for the inversion solvers.
# create PyGIMLi's ERT manager
ert_mgr = ert_manager(pg_data)
2.4. Inversion mesh#
The inversion can use a different mesh and the mesh to be used should know nothing about the mesh that was designed based on the true model. Here we first use a triangular mesh for the inversion, which makes the problem underdetermined.
inv_mesh = inversion_mesh(ert_mgr)
ax = pygimli.show(inv_mesh, showMesh=True, markers=False, colorBar=False)
ax[0].set_title("Mesh used for inversion")
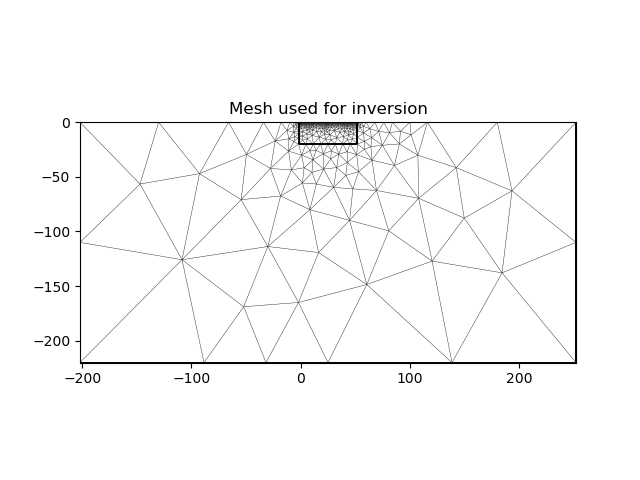
Text(0.5, 1.0, 'Mesh used for inversion')
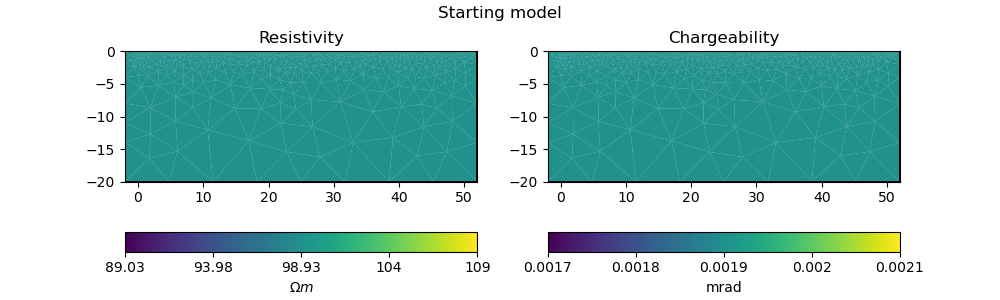
2.5. Forward operator, regularization matrix#
With the inversion mesh created, we now define a starting model, forward operator and weighting matrix for regularization using PyGIMLi.
Our model will be in log space when we perform inversion (for numerical stability purposes).
# PyGIMLi's forward operator (ERTModelling)
forward_oprt = ert_forward_operator(ert_mgr, scheme, inv_mesh)
# extract regularization matrix
Wm = reg_matrix(forward_oprt)
# initialise a starting model for inversion
start_model, start_model_log, start_model_log_real = starting_model(pg_data, ert_mgr.paraDomain)
plot_model(ert_mgr.paraDomain, start_model, "Starting model")
2.6. Utility functions to pass to CoFI#
CoFI and other inference packages require a set of functions that provide the misfit, the jacobian the residual within the case of scipy standardised interfaces. All these functions are defined below as additional utility functions, so feel free to read them into details if you want to understand more. These functions are:
get_responseget_jacobianget_residualsget_data_misfitget_regularizationget_gradientget_hessian
# Utility Functions (additional)
def _ensure_numpy(model):
if "torch.Tensor" in str(type(model)):
model = model.cpu().detach().numpy()
return model
# model_log_complex -> data_log_complex
def get_response(model_log_complex, fop):
model_complex = np.exp(model_log_complex)
model_real = complex_to_real(model_complex)
model_real = _ensure_numpy(model_real)
data_real = np.array(fop.response(model_real))
data_complex = complex_from_real(data_real)
data_log_complex = np.log(data_complex)
return data_log_complex
# model_log_complex -> J_log_log_complex
def get_jacobian(model_log_complex, fop):
model_complex = np.exp(model_log_complex)
model_real = complex_to_real(model_complex)
model_real = _ensure_numpy(model_real)
J_block = fop.createJacobian(model_real)
J_real = np.array(J_block.mat(0))
J_imag = np.array(J_block.mat(1))
J_complex = J_real + 1j * J_imag
data_log_complex = get_response(model_log_complex, fop)
data_complex = np.exp(data_log_complex)
J_log_log_complex = J_complex / data_complex[:,np.newaxis] * model_complex[np.newaxis,:]
return J_log_log_complex
# model_log_complex -> res_data_log_complex
def get_residuals(model_log_complex, data_log_complex, fop):
synth_data_log_complex = get_response(model_log_complex, fop)
return data_log_complex - synth_data_log_complex
# model_log_real -> obj_log_real
def get_objective(model_log_real, data_log_complex, fop, lamda, Wm):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate data misfit
res_log_complex = get_residuals(model_log_complex, data_log_complex, fop)
data_misfit = res_log_complex.conj().dot(res_log_complex)
# calculate regularization term
weighted_model_log_real = Wm.dot(model_log_complex)
reg = lamda * weighted_model_log_real.conj().dot(weighted_model_log_real)
# sum up
result = np.abs(data_misfit + reg)
return result
# model_log_real -> grad_log_real
def get_gradient(model_log_real, data_log_complex, fop, lamda, Wm):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate gradient for data misfit
res = get_residuals(model_log_complex, data_log_complex, fop)
jac = get_jacobian(model_log_complex, fop)
data_misfit_grad = - jac.conj().T.dot(res)
# calculate gradient for regularization term
reg_grad = lamda * Wm.T.dot(Wm).dot(model_log_complex)
# sum up
grad_complex = data_misfit_grad + reg_grad
grad_real = complex_to_real(grad_complex)
return grad_real
# model_log_real -> hess_log_real
def get_hessian(model_log_real, data_log_complex, fop, lamda, Wm):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate hessian for data misfit
res = get_residuals(model_log_complex, data_log_complex, fop)
jac = get_jacobian(model_log_complex, fop)
data_misfit_hessian = jac.conj().T.dot(jac)
# calculate hessian for regularization term
reg_hessian = lamda * Wm.T.dot(Wm)
# sum up
hessian_complex = data_misfit_hessian + reg_hessian
nparams = len(model_log_complex)
hessian_real = np.zeros((2*nparams, 2*nparams))
hessian_real[:nparams,:nparams] = np.real(hessian_complex)
hessian_real[:nparams,nparams:] = -np.imag(hessian_complex)
hessian_real[nparams:,:nparams] = np.imag(hessian_complex)
hessian_real[nparams:,nparams:] = np.real(hessian_complex)
return hessian_real
With all the above forward operations set up with PyGIMLi, we now define
the problem in cofi by setting the problem information for a
BaseProblem object.
# hyperparameters
lamda=0.001
# CoFI - define BaseProblem
dcip_problem = cofi.BaseProblem()
dcip_problem.name = "DC-IP defined through PyGIMLi"
dcip_problem.set_objective(get_objective, args=[data_log_complex, forward_oprt, lamda, Wm])
dcip_problem.set_gradient(get_gradient, args=[data_log_complex, forward_oprt, lamda, Wm])
dcip_problem.set_hessian(get_hessian, args=[data_log_complex, forward_oprt, lamda, Wm])
dcip_problem.set_initial_model(start_model_log_real)
3. Define the inversion options and run#
3.1. SciPy’s optimizer (trust-ncg)#
dcip_problem.suggest_tools();
Based on what you've provided so far, here are possible tools:
{
"optimization": [
"scipy.optimize.minimize",
"torch.optim"
],
"matrix solvers": [
"cofi.simple_newton"
],
"sampling": [
"bayesbay",
"neighpy"
]
}
{'optimization': ['scipy.optimize.minimize', 'torch.optim'], 'matrix solvers': ['cofi.simple_newton'], 'sampling': ['bayesbay', 'neighpy']}
inv_options_scipy = cofi.InversionOptions()
inv_options_scipy.set_tool("scipy.optimize.minimize")
inv_options_scipy.set_params(method="trust-ncg", options={"maxiter":5})
inv_scipy = cofi.Inversion(dcip_problem, inv_options_scipy)
inv_result_scipy = inv_scipy.run()
print(f"\nSolver message: {inv_result_scipy.message}")
Solver message: Maximum number of iterations has been exceeded.
model_scipy = np.exp(complex_from_real(inv_result_scipy.model))
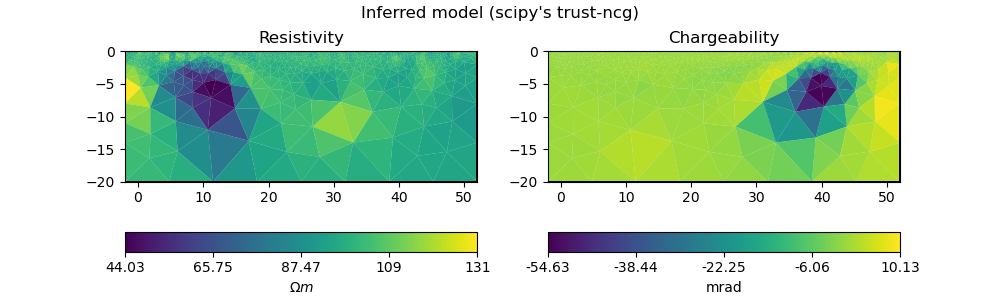
plot_model(ert_mgr.paraDomain, model_scipy, "Inferred model (scipy's trust-ncg)")
synth_data_scipy = np.exp(get_response(np.log(model_scipy), forward_oprt))
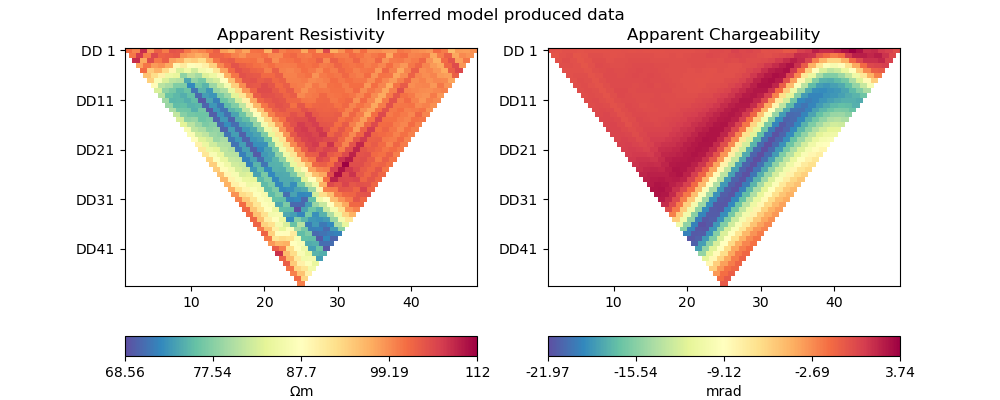
plot_data(pg_data, synth_data_scipy, "Inferred model produced data")
3.2. PyTorch’s optimizer (RAdam)#
inv_options_torch = cofi.InversionOptions()
inv_options_torch.set_tool("torch.optim")
inv_options_torch.set_params(algorithm="RAdam", lr=0.05, num_iterations=20)
inv_torch = cofi.Inversion(dcip_problem, inv_options_torch)
inv_result_torch = inv_torch.run()
Iteration #0, objective value: 40.45486222394138
Iteration #1, objective value: 32.66647384877092
Iteration #2, objective value: 27.255098186704533
Iteration #3, objective value: 23.616898709394736
Iteration #4, objective value: 21.250039852018595
Iteration #5, objective value: 19.754375173835047
Iteration #6, objective value: 19.664967415508343
Iteration #7, objective value: 19.546714992374785
Iteration #8, objective value: 19.404993594348355
Iteration #9, objective value: 19.244973185523822
Iteration #10, objective value: 19.07158854847276
Iteration #11, objective value: 18.888839174555393
Iteration #12, objective value: 18.699874814641333
Iteration #13, objective value: 18.508109556512483
Iteration #14, objective value: 18.317770581716896
Iteration #15, objective value: 18.13326509047591
Iteration #16, objective value: 17.95822138072281
Iteration #17, objective value: 17.79495112868809
Iteration #18, objective value: 17.644666903626334
Iteration #19, objective value: 17.50809850311941
model_torch = np.exp(complex_from_real(inv_result_torch.model))
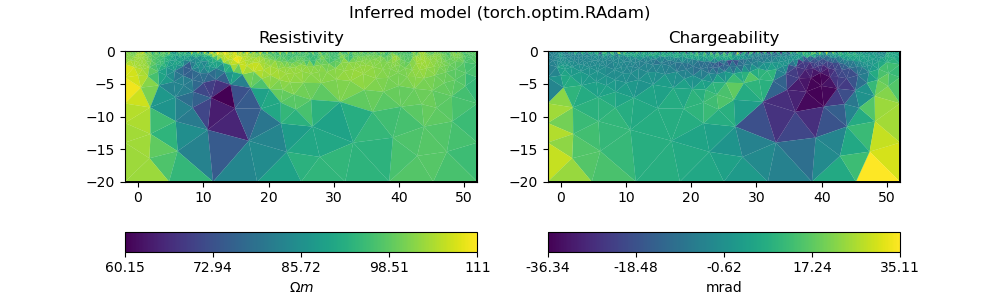
plot_model(ert_mgr.paraDomain, model_torch, "Inferred model (torch.optim.RAdam)")
synth_data_torch = np.exp(get_response(np.log(model_torch), forward_oprt))
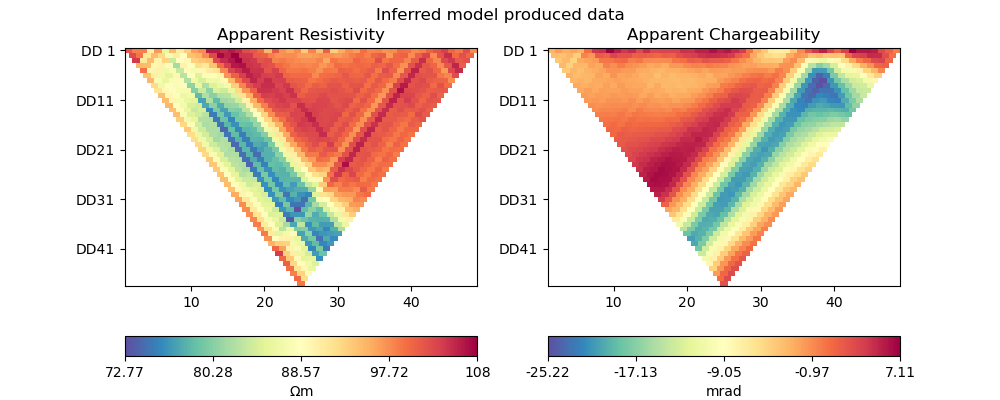
plot_data(pg_data, synth_data_torch, "Inferred model produced data")
Watermark#
watermark_list = ["cofi", "numpy", "scipy", "pygimli", "torch", "matplotlib"]
for pkg in watermark_list:
pkg_var = __import__(pkg)
print(pkg, getattr(pkg_var, "__version__"))
cofi 0.2.7
numpy 1.24.4
scipy 1.12.0
pygimli 1.4.6
torch 2.1.2.post101
matplotlib 3.8.3
sphinx_gallery_thumbnail_number = -1
Total running time of the script: (2 minutes 57.844 seconds)