Note
Go to the end to download the full example code
Century DCIP Inversion with a Triangular Mesh#
If you are running this notebook locally, make sure you’ve followed steps here to set up the environment. (This environment.yml file specifies a list of packages required to run the notebooks)
Motivation#
The Century Deposit is a zinc-lead-silver deposit in the Mt Isa region in Queensland Australia and UBC 2D DCIP inversion results have been published by Mutton, 2000 and reproduced with SimPEG. It provides an excellent test case to verify if CoFI can indeed act as a glue between forward solvers and inverse solvers and be applied to real data. Figure 1 from Mutton, 2000 provides a map of the location and geological setting for the Century deposit.
A detailed descrtiption of the geological setting is available here and Mutton, 2000 also provide the cross-section for the survey line 46800mE, which we will invert in the following.
What we are interested in is delineating the mineralised units by using
the DCIP (Direct Current, Induced Polarization) solver implemented in
PyGIMLi together with the cofi
solvers.
Some background information around how a DCIP inversion using complex numbers to express resistivity and chargeability can be implemented using CoFI is given in the synthetic example notebook. While PyGIMLi allows us to use a triangular mesh which can be adanvatengous when compared with a rectilinear mesh, it also requires the data and model to be expressed as frequency domain measurements, that is as complex numbers where the real part represents the resistivity and the phase angle the chargeability. There are several ways to capture/express chargeability and SimPEG uses apparent chargeabilities \(\mathrm{M}\). Thus prior to inversion we will convert them using the following rule of thumb \(0.1 M \approx70 \mathrm{mrad}\).
References#
Martin, T., Günther, T., Orozco, A. F., & Dahlin, T. (2020). Evaluation of spectral induced polarization field measurements in time and frequency domain. Journal of Applied Geophysics, 180. https://doi.org/10.1016/j.jappgeo.2020.104141
Mutton, A. J. (2000). The application of geophysics during evaluation of the Century zinc deposit. Geophysics, 65(6), 1946–1960. https://doi.org/10.1190/1.1444878
1. Set up environment#
We’ll do the following: 1. Install PyGIMLi (if on CoLab) 2. Download processed dataset (if on CoLab) 3. Import modules
# -------------------------------------------------------- #
# #
# Uncomment below to set up environment on "colab" #
# #
# -------------------------------------------------------- #
# !pip install -U cofi
# !pip install -q condacolab
# import condacolab
# condacolab.install()
# !mamba install -c gimli pygimli=1.3
# -------------------------------------------------------- #
# #
# Uncomment below to set up environment on "colab" #
# #
# -------------------------------------------------------- #
# !git clone https://github.com/inlab-geo/cofi-examples.git
# %cd cofi-examples/examples/pygimli_dcip
We will need the following packages:
osto list and load datasetnumpyfor matrices and matrix-related functionsmatplotlibfor plottingpygimlifor forward modelling of the problemcofifor accessing different inference solvers
import os
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
import pygimli
import cofi
2. Load the data#
We will need to download the preprocessed dataset first. This notebook century_data_preprocessing.ipynb contains the code for data preprocessing.
data_base_path = "../../data/century_dcip"
dcip_data = np.loadtxt(f"{data_base_path}/century_dcip_data.txt")
PyGIMLi expresses chargeability in \(\mathrm{radians}\) and we convert the apparent chargeabilites as we load the data.
a_locs = dcip_data[:,0]
b_locs = dcip_data[:,1]
m_locs = dcip_data[:,2]
n_locs = dcip_data[:,3]
dc_obs = dcip_data[:,4]
dc_err = dcip_data[:,5]
ip_obs = dcip_data[:,6]*0.7 # https://gpg.geosci.xyz/content/induced_polarization/induced_polarization_data.html
ip_err = dcip_data[:,7]*0.7
geo_factors = dcip_data[:,8]
location_start = np.min(a_locs)
location_stop = np.max(n_locs)
location_interval = m_locs[1] - m_locs[0]
location_num = int((location_stop - location_start) / location_interval + 1)
location_start, location_stop, location_interval, location_num
(26000.0, 29200.0, 100.0, 33)
def load_leapfrog_geologic_section(filename=f"{data_base_path}/century_geologic_section.csv"):
"""
Load the geologic cross section.
"""
fid = open(filename, 'r')
lines = fid.readlines()
data = []
data_tmp = []
for line in lines[2:]:
line_data = (line.split(',')[:3])
if 'End' in line:
data.append(np.vstack(data_tmp)[:,[0, 2]])
data_tmp = []
else:
data_tmp.append(np.array(line_data, dtype=float))
return data
geologic_section = load_leapfrog_geologic_section()
3. Utility wrappers to PyGIMLi functions#
Below we define a set of utility functions that help define the problem, generating data and making plots. Feel free to skip reading the details of these utility functions and come back later if you want.
def rho_phi_to_complex(rho, phi): # rho * e^(phi * i)
return pygimli.utils.toComplex(rho, phi)
def rho_phi_from_complex(complx): # |complx|, arctan(complx.imag, complx.real)
return np.abs(complx), np.arctan2(complx.imag, complx.real)
def complex_to_real(complx): # complx vector of size n -> size 2n
return pygimli.utils.squeezeComplex(complx)
def complex_from_real(real): # real vector of size n -> size n/2
return pygimli.utils.toComplex(real)
# inversion mesh bound
x_inv_start = location_start - 200
x_inv_stop = location_stop + 200
y_inv_start = -400
y_inv_stop = 0
# PyGIMLi DataContainerERT
def pygimli_data(a_locs, b_locs, m_locs, n_locs, dc_obs, dc_err, ip_obs, ip_err):
# --- create empty data container object ---
pg_data = pygimli.DataContainerERT()
# create sensor locations
for sensor in np.linspace(location_start, location_stop, location_num):
pg_data.createSensor((sensor, 0.0, 0.0))
# --- add indices for data points ---
locs_sources = np.vstack((a_locs, b_locs)).T
locs_receivers = np.vstack((m_locs, n_locs)).T
for i in range(len(locs_sources)):
src = locs_sources[i]
src_idx = (src - location_start) / location_interval
rec = locs_receivers[i]
rec_idx = (rec - location_start) / location_interval
pg_data.createFourPointData(i, src_idx[0], src_idx[1], rec_idx[0], rec_idx[1])
# --- fill in the observed data and error estimation ---
pg_data["rhoa"] = dc_obs
pg_data["err"] = dc_err
pg_data["phia"] = -ip_obs/1000. # PyGIMLi accepts radians (instead of milliradians) for forward modelling
pg_data["iperr"] = ip_err/1000.
# --- create geometric factor k ---
pg_data["k"] = pygimli.physics.ert.createGeometricFactors(pg_data, numerical=True)
# --- generate data vals and diag vals of covariance inv matrix in log complex space ---
data_complex = rho_phi_to_complex(pg_data["rhoa"].array(), pg_data["phia"].array())
data_log_complex = np.log(data_complex)
dc_err_log = np.log(pg_data["err"])
ip_err_log = np.log(pg_data["iperr"])
m_err = rho_phi_to_complex(1/dc_err_log, 1/ip_err_log)
Wd = np.diag(m_err)
Cd_inv = Wd.conj().dot(Wd)
return pg_data, data_log_complex, Cd_inv
# PyGIMLi ert.ERTManager
def ert_manager(pg_data, verbose=False):
return pygimli.physics.ert.ERTManager(pg_data, verbose=verbose, useBert=True)
# mesh used for inversion
def inversion_mesh(ert_mgr):
inv_mesh = ert_mgr.createMesh(ert_mgr.data)
inv_mesh = inv_mesh.createH2()
ert_mgr.setMesh(inv_mesh)
print("model size", ert_mgr.paraDomain.cellCount())
return inv_mesh
# mesh used for the original paper
def inversion_mesh_ubc(ert_mgr):
mesh_ubc = pygimli.meshtools.readMeshIO(f"{data_base_path}/century_mesh.vtk")
print("model size", mesh_ubc.cellCount())
ert_mgr.setMesh(mesh_ubc)
return mesh_ubc
# PyGIMLi ert.ERTModelling
def ert_forward_operator(ert_mgr, pg_data, inv_mesh):
forward_oprt = ert_mgr.fop
forward_oprt.setComplex(True)
forward_oprt.setData(pg_data)
forward_oprt.setMesh(inv_mesh, ignoreRegionManager=True)
return forward_oprt
# regularization matrix
def reg_matrix(forward_oprt, inv_mesh):
region_manager = forward_oprt.regionManager()
region_manager.setConstraintType(2)
region_manager.setMesh(inv_mesh)
Wm = pygimli.matrix.SparseMapMatrix()
region_manager.fillConstraints(Wm)
Wm = pygimli.utils.sparseMatrix2coo(Wm)
return Wm
def starting_model(data, inv_mesh, rho_val=None, phi_val=None):
rho_start = np.median(data["rhoa"]) if rho_val is None else rho_val
phi_start = np.median(data["phia"]) if phi_val is None else phi_val
start_model_val = rho_phi_to_complex(rho_start, phi_start)
start_model_complex = np.ones(inv_mesh.cellCount()) * start_model_val
start_model_log_complex = np.log(start_model_complex)
start_model_log_real = complex_to_real(start_model_log_complex)
return start_model_complex, start_model_log_complex, start_model_log_real
def reference_dc_model():
return np.loadtxt(f"{data_base_path}/century_dc_model.txt")
def reference_ip_model():
return -np.loadtxt(f"{data_base_path}/century_ip_model.txt") * 0.7 / 1000
# initialise model to have same resistivities as the original inversion result
def starting_model_ref(ert_mgr):
dc_model_ref = np.loadtxt(f"{data_base_path}/century_dc_model.txt")
assert ert_mgr.paraDomain.cellCount() == len(dc_model_ref), \
"mesh cell count has to match century reference model length"
return starting_model(ert_mgr, rhoa_val=dc_model_ref)
Note: We lifted out the plotting of colorbars only for Colab compatibility.
resistivity_label = r"$\Omega m$"
chargeability_label = r"mrad"
def plot_geologic_section(geologic_section, ax):
for data in geologic_section:
ax.plot(data[:,0], data[:,1], 'k--', alpha=0.5)
def plot_colorbar(ax, cMin, cMax, label, orientation="horizontal"):
norm = mpl.colors.Normalize(cMin, cMax)
sm = plt.cm.ScalarMappable(norm=norm)
cb = plt.colorbar(sm, orientation=orientation, ax=ax)
cb.set_label(label)
cb.set_ticks(np.linspace(cMin, cMax, 5, endpoint=True))
def plot_model(mesh, model_complex, title):
rho, phi = rho_phi_from_complex(model_complex)
fig, axes = plt.subplots(2,1,figsize=(12,5))
pygimli.show(mesh, data=rho, label=resistivity_label, ax=axes[0], colorBar=False)
axes[0].set_xlim(x_inv_start, x_inv_stop)
axes[0].set_ylim(y_inv_start, y_inv_stop)
axes[0].set_title("Resistivity")
plot_colorbar(axes[0], 136, 170, resistivity_label)
pygimli.show(mesh, data=phi * 1000, label=chargeability_label, cMin=-4.76, cMax=-4, ax=axes[1], colorBar=False)
axes[1].set_xlim(x_inv_start, x_inv_stop)
axes[1].set_ylim(y_inv_start, y_inv_stop)
axes[1].set_title("Chargeability")
plot_colorbar(axes[1], -4.76, -4, chargeability_label)
if title != "Starting model":
plot_geologic_section(geologic_section, axes[0])
plot_geologic_section(geologic_section, axes[1])
fig.suptitle(title)
def plot_data(pg_data, data_complex, title):
rho, phi = rho_phi_from_complex(data_complex)
fig, axes = plt.subplots(1,2,figsize=(10,4))
# pygimli.physics.ert.showERTData(pg_data, vals=rho, label=resistivity_label, ax=axes[0], colorBar=False)
pygimli.physics.ert.showERTData(pg_data, vals=rho, ax=axes[0], colorBar=False)
axes[0].set_title("Apparent Resistivity")
plot_colorbar(axes[0], np.min(rho), np.max(rho), resistivity_label)
pygimli.physics.ert.showERTData(pg_data, vals=phi*1000, ax=axes[1], colorBar=False)
# pygimli.physics.ert.showERTData(pg_data, vals=phi*1000, label=chargeability_label, ax=axes[1], colorBar=False)
axes[1].set_title("Apparent Chargeability")
plot_colorbar(axes[1], np.min(phi*1000), np.max(phi*1000), chargeability_label)
fig.suptitle(title)
def plot_mesh(mesh, title="Mesh used for inversion"):
_, ax = plt.subplots(1, 1)
pygimli.show(mesh, showMesh=True, markers=False, colorBar=False, ax=ax)
ax.set_title(title)
ax.set_xlabel("Northing (m)")
ax.set_ylabel("Elevation (m)")
def plot_comparison(mesh1, model1, title1, mesh2, model2, title2, rho_min, rho_max, phi_min, phi_max):
rho1, phi1 = rho_phi_from_complex(model1)
rho2, phi2 = rho_phi_from_complex(model2)
fig, axes = plt.subplots(4, 1, figsize=(10,12))
pygimli.show(mesh1, data=rho1, label=resistivity_label, ax=axes[0], colorBar=False)
axes[0].set_xlim(x_inv_start, x_inv_stop)
axes[0].set_ylim(y_inv_start, y_inv_stop)
axes[0].set_title(f"{title1} - Resistivity")
plot_colorbar(axes[0], rho_min, rho_max, resistivity_label)
plot_geologic_section(geologic_section, axes[0])
pygimli.show(mesh2, data=rho2, label=resistivity_label, ax=axes[1], cMin=rho_min, cMax=rho_max, colorBar=False)
axes[1].set_xlim(x_inv_start, x_inv_stop)
axes[1].set_ylim(y_inv_start, y_inv_stop)
axes[1].set_title(f"{title2} - Resistivity")
plot_colorbar(axes[1], rho_min, rho_max, resistivity_label)
plot_geologic_section(geologic_section, axes[1])
pygimli.show(mesh1, data=phi1 * 1000, label=chargeability_label, ax=axes[2], colorBar=False)
axes[2].set_xlim(x_inv_start, x_inv_stop)
axes[2].set_ylim(y_inv_start, y_inv_stop)
axes[2].set_title(f"{title1} - Chargeability")
plot_colorbar(axes[2], phi_min*1000, phi_max*1000, chargeability_label)
plot_geologic_section(geologic_section, axes[2])
pygimli.show(mesh2, data=phi2 * 1000, label=chargeability_label, ax=axes[3], cMin=phi_min*1000, cMax=phi_max*1000, colorBar=False)
axes[3].set_xlim(x_inv_start, x_inv_stop)
axes[3].set_ylim(y_inv_start, y_inv_stop)
axes[3].set_title(f"{title2} - Chargeability")
plot_colorbar(axes[3], phi_min*1000, phi_max*1000, chargeability_label)
plot_geologic_section(geologic_section, axes[3])
4. PyGIMLi problem setup#
17/04/24 - 16:10:38 - pyGIMLi - INFO - Create default mesh for geometric factor calculation.
17/04/24 - 16:10:40 - pyGIMLi - INFO - Save RVector binary
17/04/24 - 16:10:40 - pyGIMLi - INFO - Cache stored: /home/jiawen/.cache/pygimli/303729043524732118
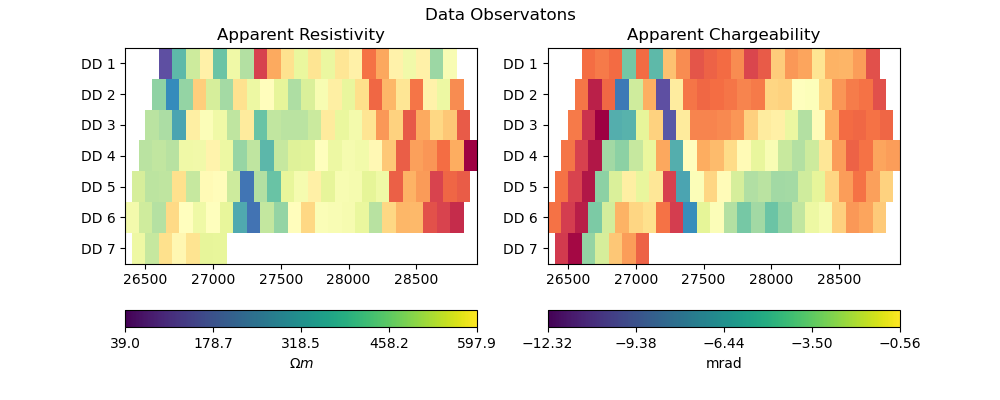
Data: Sensors: 33 data: 151, nonzero entries: ['a', 'b', 'err', 'iperr', 'k', 'm', 'n', 'phia', 'rhoa', 'valid']
# this cell needs to be run twice in order to work well
plot_data(pg_data, np.exp(data_log_complex), "Data Observatons")
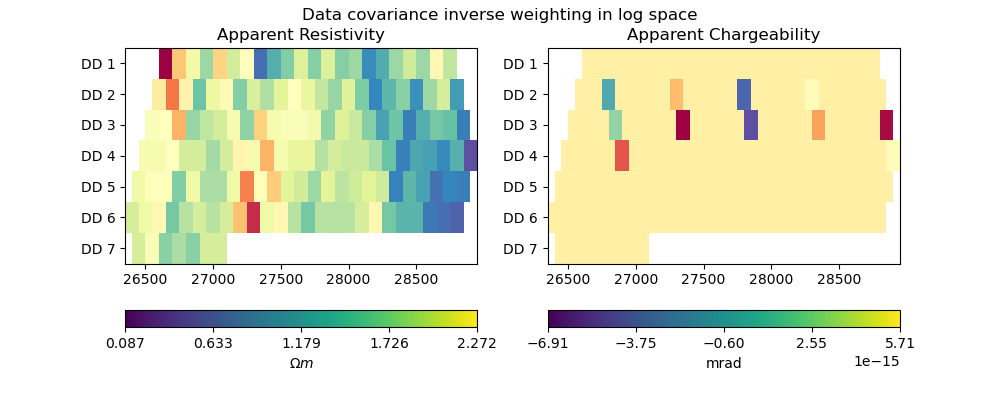
plot_data(pg_data, np.diag(Cd_inv), "Data covariance inverse weighting in log space")
ert_mgr = ert_manager(pg_data)
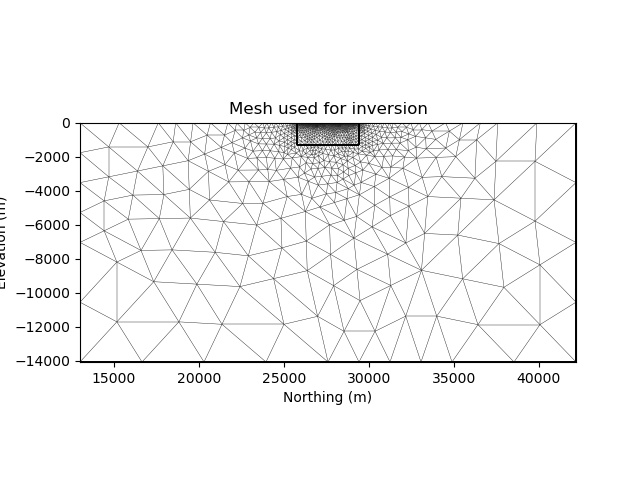
inv_mesh = inversion_mesh(ert_mgr)
# inv_mesh = inversion_mesh_ubc(ert_mgr)
plot_mesh(inv_mesh)
17/04/24 - 16:10:40 - pyGIMLi - INFO - Found 2 regions.
17/04/24 - 16:10:40 - pyGIMLi - INFO - (ERTModelling) Region with smallest marker (1) set to background.
17/04/24 - 16:10:40 - pyGIMLi - INFO - Found 2 regions.
17/04/24 - 16:10:40 - pyGIMLi - INFO - (ERTModelling) Region with smallest marker (1) set to background.
17/04/24 - 16:10:40 - pyGIMLi - INFO - Creating forward mesh from region infos.
17/04/24 - 16:10:40 - pyGIMLi - INFO - Creating refined mesh (H2) to solve forward task.
17/04/24 - 16:10:40 - pyGIMLi - INFO - Mesh for forward task: Mesh: Nodes: 5657 Cells: 10912 Boundaries: 8384
model size 1912
forward_oprt = ert_forward_operator(ert_mgr, pg_data, ert_mgr.paraDomain)
Wm = reg_matrix(forward_oprt, ert_mgr.paraDomain)
17/04/24 - 16:10:41 - Core - INFO - More than 50 regions, so we assume single regions only.
17/04/24 - 16:10:41 - Core - INFO - Applying *:* interregion constraints.
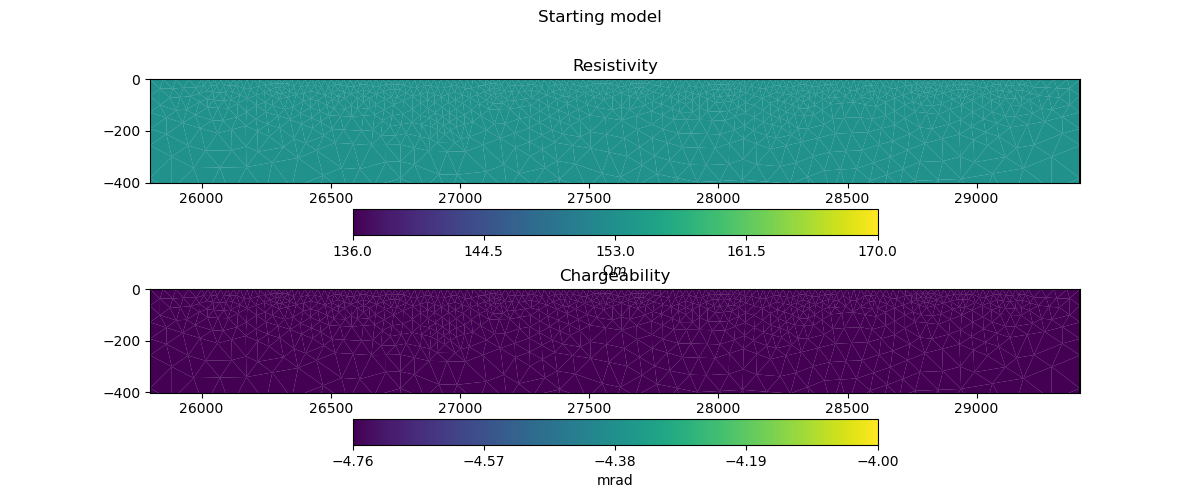
start_model_complex, start_model_log_complex, start_model_log_real = starting_model(pg_data, ert_mgr.paraDomain)
plot_model(ert_mgr.paraDomain, start_model_complex, "Starting model")
5. Create utility functions to pass to CoFI#
CoFI and other inference packages require a set of functions that provide the misfit, the jacobian the residual within the case of scipy standardised interfaces. All these functions are defined below as additional utility functions, so feel free to read them into details if you want to understand more. These functions are:
get_responseget_jacobianget_residualsget_data_misfitget_regularizationget_gradientget_hessian
# Utility Functions (additional)
def _ensure_numpy(model):
if "torch.Tensor" in str(type(model)):
model = model.cpu().detach().numpy()
return model
# model_log_complex -> data_log_complex
def get_response(model_log_complex, fop):
model_complex = np.exp(model_log_complex)
model_real = complex_to_real(model_complex)
model_real = _ensure_numpy(model_real)
data_real = np.array(fop.response(model_real))
data_complex = complex_from_real(data_real)
data_log_complex = np.log(data_complex)
return data_log_complex
# model_log_complex -> J_log_log_complex
def get_jacobian(model_log_complex, fop):
model_complex = np.exp(model_log_complex)
model_real = complex_to_real(model_complex)
model_real = _ensure_numpy(model_real)
J_block = fop.createJacobian(model_real)
J_real = np.array(J_block.mat(0))
J_imag = np.array(J_block.mat(1))
J_complex = J_real + 1j * J_imag
data_log_complex = get_response(model_log_complex, fop)
data_complex = np.exp(data_log_complex)
J_log_log_complex = J_complex / data_complex[:,np.newaxis] * model_complex[np.newaxis,:]
return J_log_log_complex
# model_log_complex -> res_data_log_complex
def get_residuals(model_log_complex, data_log_complex, fop):
synth_data_log_complex = get_response(model_log_complex, fop)
return data_log_complex - synth_data_log_complex
# model_log_real -> obj_log_real
def get_objective(model_log_real, data_log_complex, fop, lamda, Wm, Cd_inv):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate data misfit
res_log_complex = get_residuals(model_log_complex, data_log_complex, fop)
data_misfit = res_log_complex.conj().dot(Cd_inv).dot(res_log_complex)
# calculate regularization term
weighted_model_log_real = Wm.dot(model_log_complex)
reg = lamda * weighted_model_log_real.conj().dot(weighted_model_log_real)
# sum up
print(f"data misfit: {np.abs(data_misfit)}, reg: {np.abs(reg)}")
result = np.abs(data_misfit + reg)
return result
# model_log_real -> grad_log_real
def get_gradient(model_log_real, data_log_complex, fop, lamda, Wm, Cd_inv):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate gradient for data misfit
res = get_residuals(model_log_complex, data_log_complex, fop)
jac = get_jacobian(model_log_complex, fop)
data_misfit_grad = - jac.conj().T.dot(Cd_inv).dot(res)
# calculate gradient for regularization term
reg_grad = lamda * Wm.T.dot(Wm).dot(model_log_complex)
# sum up
grad_complex = data_misfit_grad + reg_grad
grad_real = complex_to_real(grad_complex)
return grad_real
# model_log_real -> hess_log_real
def get_hessian(model_log_real, data_log_complex, fop, lamda, Wm, Cd_inv):
# convert model_log_real into complex numbers
model_log_complex = complex_from_real(model_log_real)
# calculate hessian for data misfit
res = get_residuals(model_log_complex, data_log_complex, fop)
jac = get_jacobian(model_log_complex, fop)
data_misfit_hessian = jac.conj().T.dot(Cd_inv).dot(jac)
# calculate hessian for regularization term
reg_hessian = lamda * Wm.T.dot(Wm)
# sum up
hessian_complex = data_misfit_hessian + reg_hessian
nparams = len(model_log_complex)
hessian_real = np.zeros((2*nparams, 2*nparams))
hessian_real[:nparams,:nparams] = np.real(hessian_complex)
hessian_real[:nparams,nparams:] = -np.imag(hessian_complex)
hessian_real[nparams:,:nparams] = np.imag(hessian_complex)
hessian_real[nparams:,nparams:] = np.real(hessian_complex)
return hessian_real
# test
try:
get_response(start_model_log_real, forward_oprt)
except RuntimeError:
print("run again")
get_response(start_model_log_real, forward_oprt)
run again
# test
obj_val = get_objective(start_model_log_real, data_log_complex, forward_oprt, 0.0001, Wm, Cd_inv)
obj_val
data misfit: 16.48522124495534, reg: 0.0
16.48522124495534
# test
gradient = get_gradient(start_model_log_real, data_log_complex, forward_oprt, 0.0001, Wm, Cd_inv)
gradient.shape, gradient
((3824,), 3824 [-0.03303060426070571,...,-9.278041136645493e-06])
((3824, 3824), array([[ 5.38948398e-03, 2.13205609e-03, 2.52937620e-03, ...,
-7.74805593e-17, -7.06284584e-17, -6.98105777e-17],
[ 2.13205609e-03, 1.52260555e-03, 1.36485591e-03, ...,
-3.83824559e-17, -3.39835725e-17, -3.42444636e-17],
[ 2.52937620e-03, 1.36485591e-03, 2.07533134e-03, ...,
-4.44996868e-17, -3.91433729e-17, -3.97066535e-17],
...,
[-7.74805553e-17, -3.83824545e-17, -4.44996846e-17, ...,
3.01850225e-04, 1.83937474e-06, -9.81551500e-05],
[-7.06284549e-17, -3.39835713e-17, -3.91433709e-17, ...,
1.83937474e-06, 2.01883370e-04, -9.81077661e-05],
[-6.98105752e-17, -3.42444627e-17, -3.97066521e-17, ...,
-9.81551500e-05, -9.81077661e-05, 3.01974672e-04]]))
With all the above forward operations set up with PyGIMLi, we now define
the problem in cofi by setting the problem information for a
BaseProblem object.
# hyperparameters
lamda=0.001
# CoFI - define BaseProblem
dcip_problem = cofi.BaseProblem()
dcip_problem.name = "DC-IP defined through PyGIMLi"
dcip_problem.set_objective(get_objective, args=[data_log_complex, forward_oprt, lamda, Wm, Cd_inv])
dcip_problem.set_gradient(get_gradient, args=[data_log_complex, forward_oprt, lamda, Wm, Cd_inv])
dcip_problem.set_hessian(get_hessian, args=[data_log_complex, forward_oprt, lamda, Wm, Cd_inv])
dcip_problem.set_initial_model(start_model_log_real)
dcip_problem.suggest_tools();
Based on what you've provided so far, here are possible tools:
{
"optimization": [
"scipy.optimize.minimize",
"torch.optim"
],
"matrix solvers": [
"cofi.simple_newton"
],
"sampling": [
"bayesbay",
"neighpy"
]
}
{'optimization': ['scipy.optimize.minimize', 'torch.optim'], 'matrix solvers': ['cofi.simple_newton'], 'sampling': ['bayesbay', 'neighpy']}
6. Define the inversion options and run#
Triangular mesh solved with SciPy’s optimizer (trust-ncg)
inv_options_scipy = cofi.InversionOptions()
inv_options_scipy.set_tool("scipy.optimize.minimize")
class CallBack:
def __init__(self):
self._i = 1
def __call__(self, x):
print(f"Iteration #{self._i}, objective value: {dcip_problem.objective(x)}")
self._i += 1
inv_options_scipy.set_params(method="trust-ncg", options={"maxiter":10}, callback=CallBack())
inv_scipy = cofi.Inversion(dcip_problem, inv_options_scipy)
inv_result_scipy = inv_scipy.run()
print(f"\nSolver message: {inv_result_scipy.message}")
data misfit: 16.48522124495534, reg: 0.0
data misfit: 13.070065613429875, reg: 0.0011545790036786038
data misfit: 13.070065613429875, reg: 0.0011545790036786038
Iteration #1, objective value: 13.071220192433554
data misfit: 8.1495308290436, reg: 0.010185794288245668
data misfit: 8.1495308290436, reg: 0.010185794288245668
Iteration #2, objective value: 8.159716623331846
data misfit: 4.195195896395698, reg: 0.04204069484077912
data misfit: 4.195195896395698, reg: 0.04204069484077912
Iteration #3, objective value: 4.237236591236477
data misfit: 1.52027085528683, reg: 0.0882713073531516
data misfit: 1.52027085528683, reg: 0.0882713073531516
Iteration #4, objective value: 1.6085421626399816
data misfit: 0.2834523500151795, reg: 0.10407508740263018
data misfit: 0.2834523500151795, reg: 0.10407508740263018
Iteration #5, objective value: 0.38752743741780965
data misfit: 0.06483596934716976, reg: 0.09306755759830967
data misfit: 0.06483596934716976, reg: 0.09306755759830967
Iteration #6, objective value: 0.15790352694547943
data misfit: 0.041215627932776264, reg: 0.08547780709519273
data misfit: 0.041215627932776264, reg: 0.08547780709519273
Iteration #7, objective value: 0.12669343502796898
data misfit: 0.038970929910762835, reg: 0.07253155029938924
data misfit: 0.038970929910762835, reg: 0.07253155029938924
Iteration #8, objective value: 0.11150248021015208
data misfit: 0.022376011418235944, reg: 0.07117681923764022
data misfit: 0.022376011418235944, reg: 0.07117681923764022
Iteration #9, objective value: 0.09355283065587616
data misfit: 0.02292643614129331, reg: 0.06704505021084833
data misfit: 0.02292643614129331, reg: 0.06704505021084833
Iteration #10, objective value: 0.08997148635214164
Solver message: Maximum number of iterations has been exceeded.
model_scipy = np.exp(complex_from_real(inv_result_scipy.model))
# plot_model(inv_mesh, model_scipy, "Inferred model (scipy's trust-ncg)")
synth_data_scipy = np.exp(get_response(np.log(model_scipy), forward_oprt))
# plot_data(pg_data, synth_data_scipy, "Inferred model produced data")
model_ref_dc = reference_dc_model()
model_ref_ip = reference_ip_model()
model_ref = rho_phi_to_complex(model_ref_dc, model_ref_ip)
mesh_ref_x = np.loadtxt(f"{data_base_path}/century_mesh_nodes_x.txt")
mesh_ref_z = np.loadtxt(f"{data_base_path}/century_mesh_nodes_z.txt")
mesh_ref = pygimli.meshtools.createMesh2D(mesh_ref_x, mesh_ref_z)
plot_comparison(mesh_ref,
model_ref,
"Mutton A. J. (2000).",
ert_mgr.paraDomain,
model_scipy,
"Inference result",
np.min(model_ref_dc),
np.max(model_ref_dc),
np.min(model_ref_ip),
np.max(model_ref_ip),
)
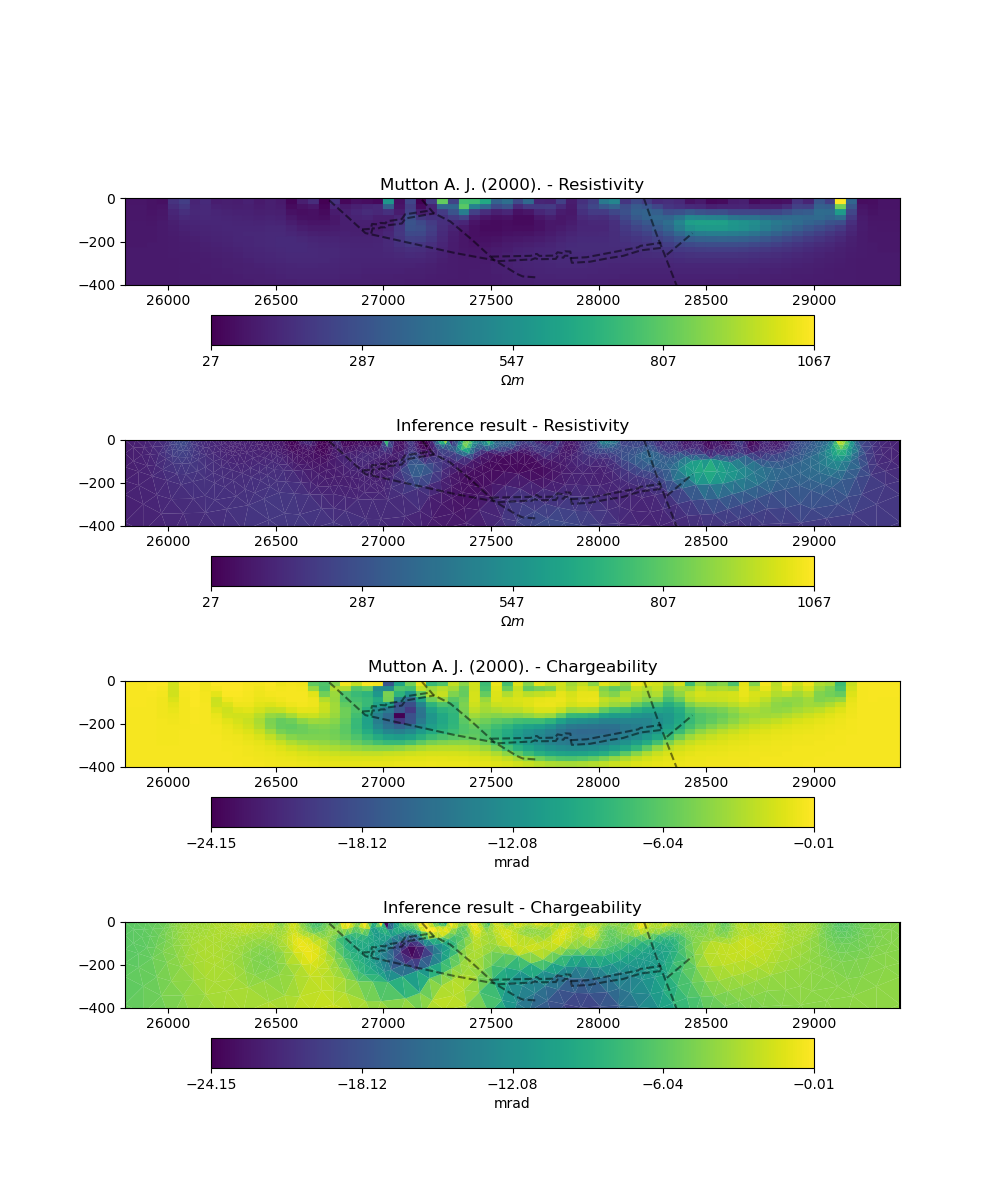
The use of an adaptive triangular mesh means that we use fewer model parameters when compared with the original example and that our mesh is reflective of the underlying physics. This speeds up the forward problem and in turn means that the inverse problem is less under-determined and a simpler regularisation (i.e. smoothing) in a single stage inversion is sufficient to obtain a result that compares favorably with the original solution.
print("Model size in the original Mutton paper:", mesh_ref.cellCount())
print("Model size of our model:", ert_mgr.paraDomain.cellCount())
Model size in the original Mutton paper: 2204
Model size of our model: 1912
Watermark#
watermark_list = ["cofi", "numpy", "scipy", "pygimli", "torch", "matplotlib"]
for pkg in watermark_list:
pkg_var = __import__(pkg)
print(pkg, getattr(pkg_var, "__version__"))
cofi 0.2.7
numpy 1.24.4
scipy 1.12.0
pygimli 1.4.6
torch 2.1.2.post101
matplotlib 3.8.3
sphinx_gallery_thumbnail_number = -1
Total running time of the script: (1 minutes 56.132 seconds)