Note
Go to the end to download the full example code
Non-linear Curve Fitting#
If you are running this notebook locally, make sure you’ve followed steps here to set up the environment. (This environment.yml file specifies a list of packages required to run the notebooks)
In this notebook, we use cofi to run a non-linear curve fitting
problem:
\[f(x)=\exp(a*x)+b\]
Import modules#
# -------------------------------------------------------- #
# #
# Uncomment below to set up environment on "colab" #
# #
# -------------------------------------------------------- #
# !pip install -U cofi
import numpy as np
import matplotlib.pyplot as plt
import arviz as az
from cofi import BaseProblem, InversionOptions, Inversion
np.random.seed(42)
Define the problem#
# Choose the "true" parameters.
a_true = 5.0
b_true = 4.0
f_true = 0.1
m_true = [a_true,b_true]
mf_true= [a_true,b_true,f_true]
# Generate some synthetic data from the model.
N = 50
x = np.sort(1 * np.random.rand(N))
yerr = 0.1 + 0.5 * np.random.rand(N)
y = my_forward(m_true,x)
y += np.abs(f_true * y) * np.random.randn(N)
y += yerr * np.random.randn(N)
plt.errorbar(x, y, yerr=yerr, fmt=".k", capsize=0)
x0 = np.linspace(0, 1, 500)
plt.plot(x0, my_forward(m_true,x0), "k", alpha=0.3, lw=3)
plt.xlim(0, 1)
plt.xlabel("x")
plt.ylabel("y");
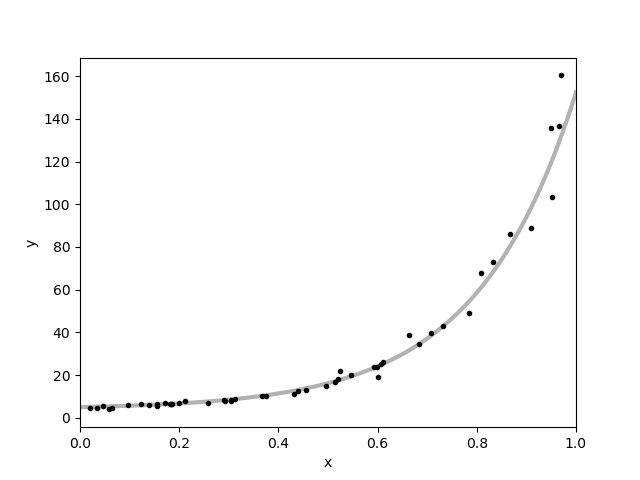
Text(38.097222222222214, 0.5, 'y')
Example 1. least squares optimizer (levenber marquardt)#
inv_options = InversionOptions()
inv_options.set_tool("scipy.optimize.least_squares")
inv_options.set_params(method="lm", max_nfev=10)
######## Run it
inv = Inversion(inv_problem, inv_options)
inv_result = inv.run()
######## Check result
print(f"The inversion result from `scipy.optimize.minimize`: {inv_result.model}\n")
inv_result.summary()
The inversion result from `scipy.optimize.minimize`: [5.06442618 3.54842172]
============================
Summary for inversion result
============================
SUCCESS
----------------------------
cost: 751.5703778228749
fun: [ 8.46834974e-02 -1.77230955e-02 -5.52853293e-01 8.89806503e-01
2.91152920e-01 -6.80792317e-01 -1.14702071e+00 -2.15801090e-01
1.82952940e-01 -5.26482030e-01 -7.76017779e-01 -5.59530381e-01
-4.95847931e-01 -4.13394792e-01 -5.36314270e-01 -1.56467760e+00
4.20608348e-01 -1.91245184e-01 -7.95757076e-02 4.30437727e-01
-1.36307871e-02 -3.20414157e-01 -3.61292253e-01 -1.97016377e-01
1.47256652e+00 1.95462598e-01 6.42560479e-01 1.17710109e+00
1.82720280e-01 -5.85651733e-01 -4.32433161e+00 -4.33451431e-01
1.59207006e-02 4.24747095e-01 5.23801008e+00 2.40244378e-01
-2.85673020e-01 -6.65912029e+00 1.06971709e+00 -1.41328842e-01
1.44236334e+00 7.70525925e+00 -4.25388813e+00 -1.75601284e+00
-1.98652707e+00 1.44619318e+01 -9.86284710e+00 2.35903628e+01
-2.98371685e-02 -2.11903105e+01]
jac: [[2.28462443e-02 1.00000000e+00]
[4.09307227e-02 1.00000000e+00]
[5.87699128e-02 1.00000000e+00]
[7.79481395e-02 1.00000000e+00]
[9.04348484e-02 1.00000000e+00]
[1.60175374e-01 1.00000000e+00]
[2.26419173e-01 1.00000000e+00]
[2.82725638e-01 1.00000000e+00]
[3.43725582e-01 1.00000000e+00]
[3.43820726e-01 1.00000000e+00]
[4.04431999e-01 1.00000000e+00]
[4.56634655e-01 1.00000000e+00]
[4.64300862e-01 1.00000000e+00]
[4.71420526e-01 1.00000000e+00]
[5.48900988e-01 1.00000000e+00]
[6.22385905e-01 1.00000000e+00]
[9.59632427e-01 1.00000000e+00]
[1.27285650e+00 1.00000000e+00]
[1.28279178e+00 1.00000000e+00]
[1.42031878e+00 1.00000000e+00]
[1.42473141e+00 1.00000000e+00]
[1.51128333e+00 1.00000000e+00]
[2.34264052e+00 1.00000000e+00]
[2.49621211e+00 1.00000000e+00]
[3.85009064e+00 1.00000000e+00]
[4.08975791e+00 1.00000000e+00]
[4.59341502e+00 1.00000000e+00]
[6.07964753e+00 1.00000000e+00]
[6.95336935e+00 1.00000000e+00]
[7.24310829e+00 1.00000000e+00]
[7.48401301e+00 1.00000000e+00]
[8.71405798e+00 1.00000000e+00]
[1.19018190e+01 1.00000000e+00]
[1.24136629e+01 1.00000000e+00]
[1.26206405e+01 1.00000000e+00]
[1.31778419e+01 1.00000000e+00]
[1.35640163e+01 1.00000000e+00]
[1.89839544e+01 1.00000000e+00]
[2.18847690e+01 1.00000000e+00]
[2.55534573e+01 1.00000000e+00]
[2.98190139e+01 1.00000000e+00]
[4.18720384e+01 1.00000000e+00]
[4.84904621e+01 1.00000000e+00]
[5.63991081e+01 1.00000000e+00]
[6.96176523e+01 1.00000000e+00]
[9.09334829e+01 1.00000000e+00]
[1.15942416e+02 1.00000000e+00]
[1.17246757e+02 1.00000000e+00]
[1.28432017e+02 1.00000000e+00]
[1.31826241e+02 1.00000000e+00]]
grad: [1.46155217e-04 9.56170254e-10]
optimality: 0.0001461552166447607
active_mask: [0 0]
nfev: 7
njev: 5
status: 2
message: `ftol` termination condition is satisfied.
model: [5.06442618 3.54842172]
Example 2. emcee#
m_min = [0,0] # lower bound for uniform prior
m_max = [10,10] # upper bound for uniform prior
def my_log_prior(m,m_min,m_max): # uniform distribution
for i in range(len(m)):
if m[i] < m_min[i] or m[i] > m_max[i]: return -np.inf
return 0.0 # model lies within bounds -> return log(1)
nwalkers = 12
ndim = 2
nsteps = 500
walkers_start = np.array([5.,4.]) + 1e-1 * np.random.randn(nwalkers, ndim)
inv_options = InversionOptions()
inv_options.set_tool("emcee")
inv_options.set_params(nwalkers=nwalkers, nsteps=nsteps, initial_state=walkers_start)
######## Run it
inv = Inversion(inv_problem, inv_options)
inv_result = inv.run()
######## Check result
print(f"The inversion result from `emcee`:")
inv_result.summary()
The inversion result from `emcee`:
============================
Summary for inversion result
============================
SUCCESS
----------------------------
sampler: <emcee.ensemble.EnsembleSampler object>
blob_names: ['log_likelihood', 'log_prior']
sampler = inv_result.sampler
az_idata = inv_result.to_arviz()
labels = ["m0", "m1"]
az.plot_trace(az_idata);
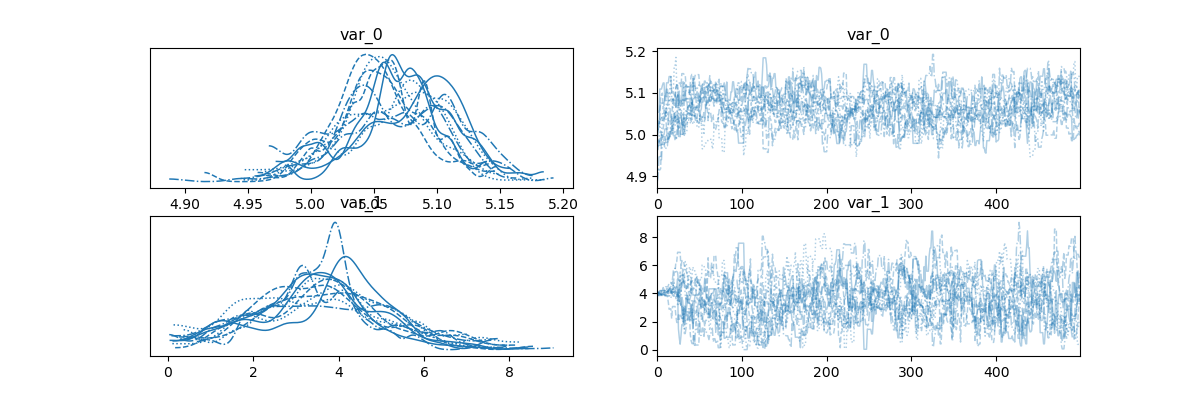
array([[<Axes: title={'center': 'var_0'}>,
<Axes: title={'center': 'var_0'}>],
[<Axes: title={'center': 'var_1'}>,
<Axes: title={'center': 'var_1'}>]], dtype=object)
_, axes = plt.subplots(2, 2, figsize=(14,10))
az.plot_pair(
az_idata.sel(draw=slice(300,None)),
marginals=True,
reference_values=dict(zip([f"var_{i}" for i in range(2)], m_true )),
ax = axes
);
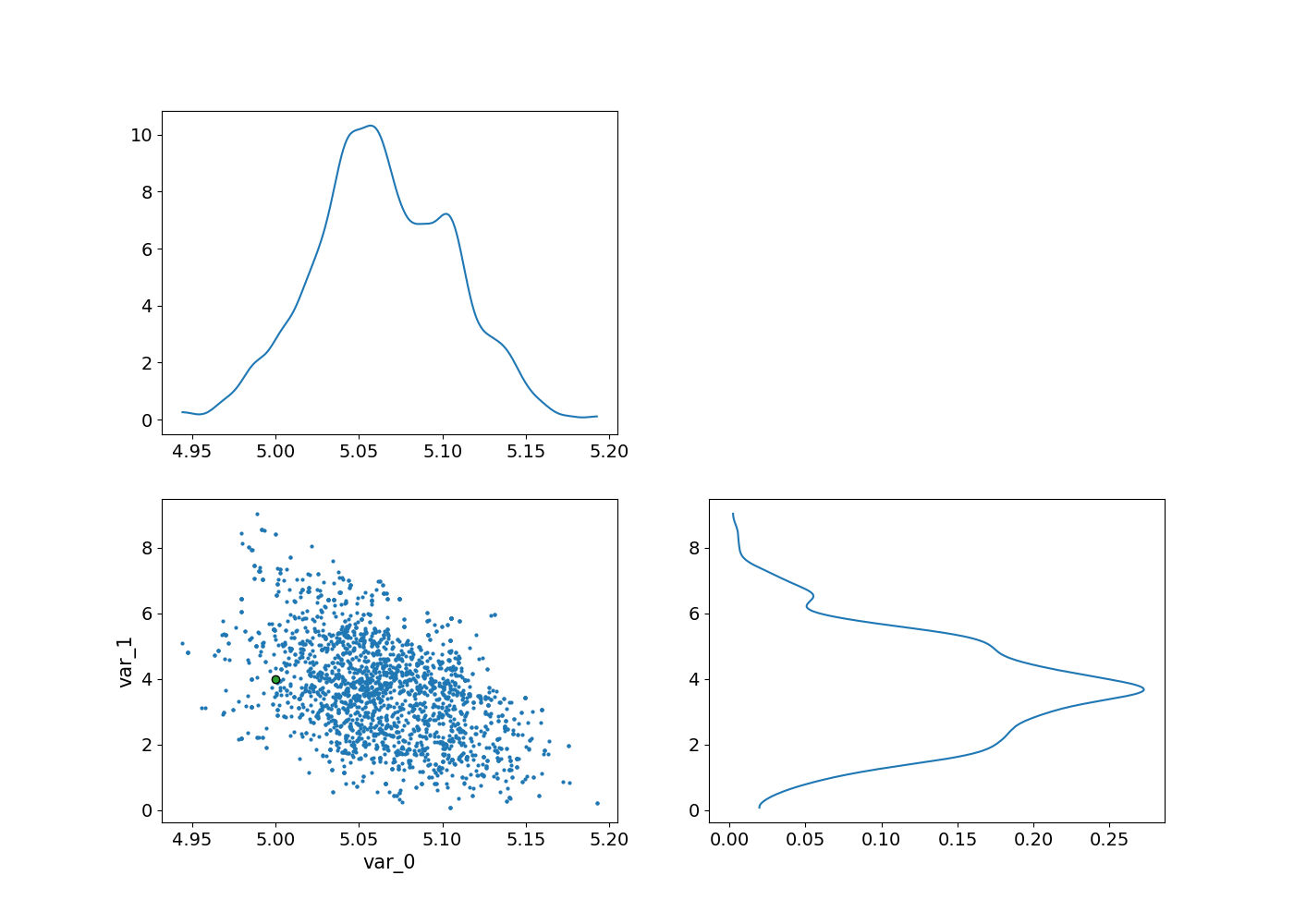
array([[<Axes: >, <Axes: >],
[<Axes: xlabel='var_0', ylabel='var_1'>, <Axes: >]], dtype=object)
flat_samples = sampler.get_chain(discard=300, thin=30, flat=True)
inds = np.random.randint(len(flat_samples), size=100) # get a random selection from posterior ensemble
_x_plot = np.linspace(0,1.0)
_y_plot = my_forward(m_true,_x_plot)
plt.figure(figsize=(12,8))
sample = flat_samples[0]
_y_synth = my_forward(sample,_x_plot)
plt.plot(_x_plot, _y_synth, color="seagreen", label="Posterior samples",alpha=0.1)
for ind in inds:
sample = flat_samples[ind]
_y_synth = my_forward(sample,_x_plot)
plt.plot(_x_plot, _y_synth, color="seagreen", alpha=0.1)
plt.plot(_x_plot, _y_plot, color="darkorange", label="true model")
plt.scatter(x, y, color="lightcoral", label="observed data")
plt.xlabel("X")
plt.ylabel("Y")
plt.legend();
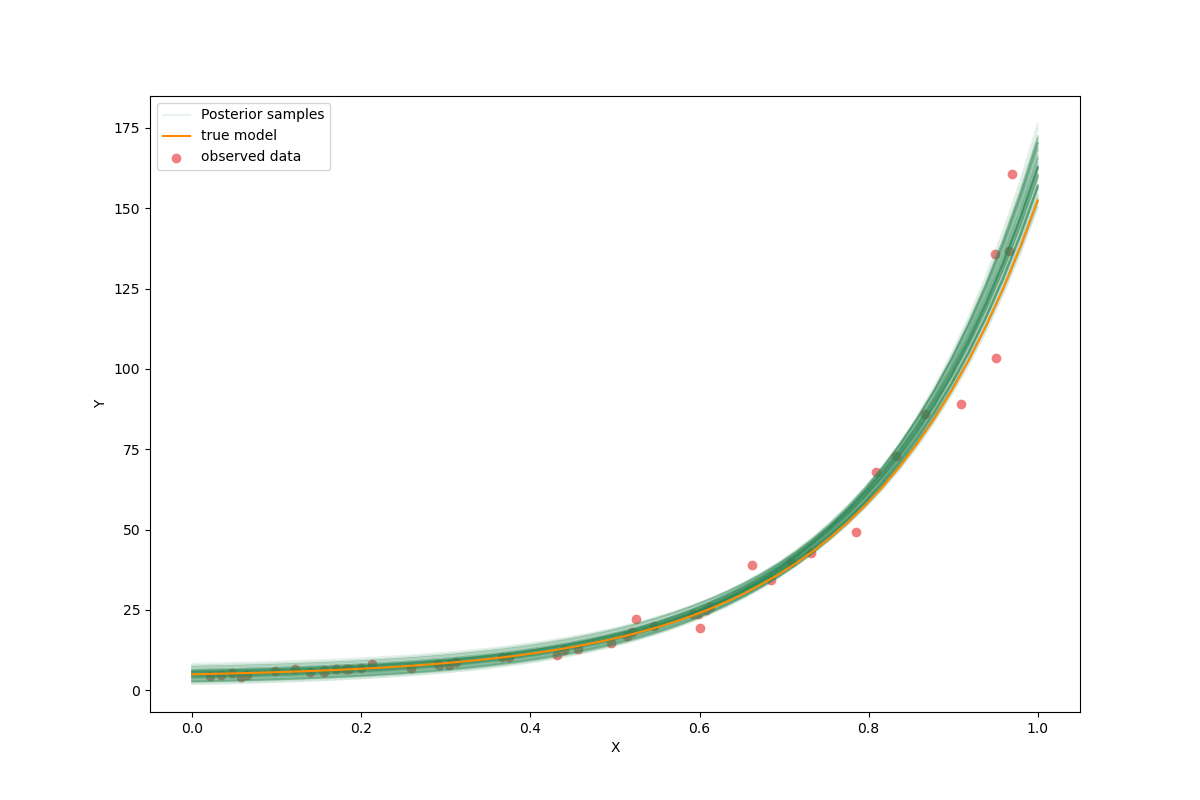
<matplotlib.legend.Legend object at 0x7f8252221b70>
Watermark#
watermark_list = ["cofi", "numpy", "scipy", "matplotlib", "emcee", "arviz"]
for pkg in watermark_list:
pkg_var = __import__(pkg)
print(pkg, getattr(pkg_var, "__version__"))
cofi 0.2.7
numpy 1.24.4
scipy 1.12.0
matplotlib 3.8.3
emcee 3.1.4
arviz 0.17.0
sphinx_gallery_thumbnail_number = -1
Total running time of the script: (0 minutes 0.879 seconds)

